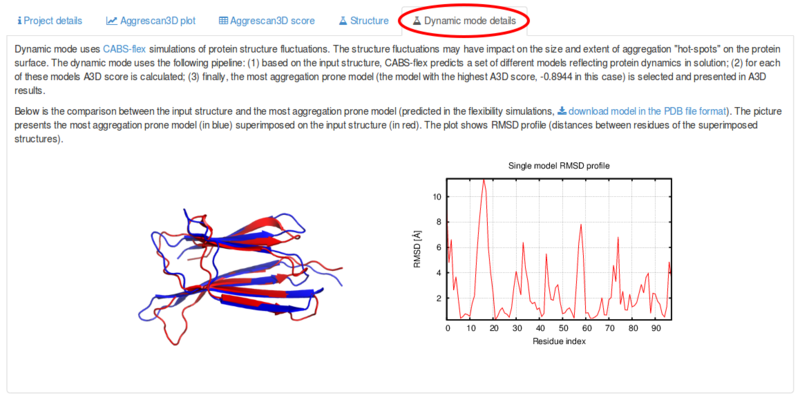
AGGRESCAN: a server for the prediction and evaluation of "hot spots" of aggregation in polypeptides | BMC Bioinformatics | Full Text

Mutational analysis of the neurexin/neuroligin complex reveals essential and regulatory components | PNAS
A3D 2.0 as a tool for the in silico redesign of more stable and soluble... | Download Scientific Diagram

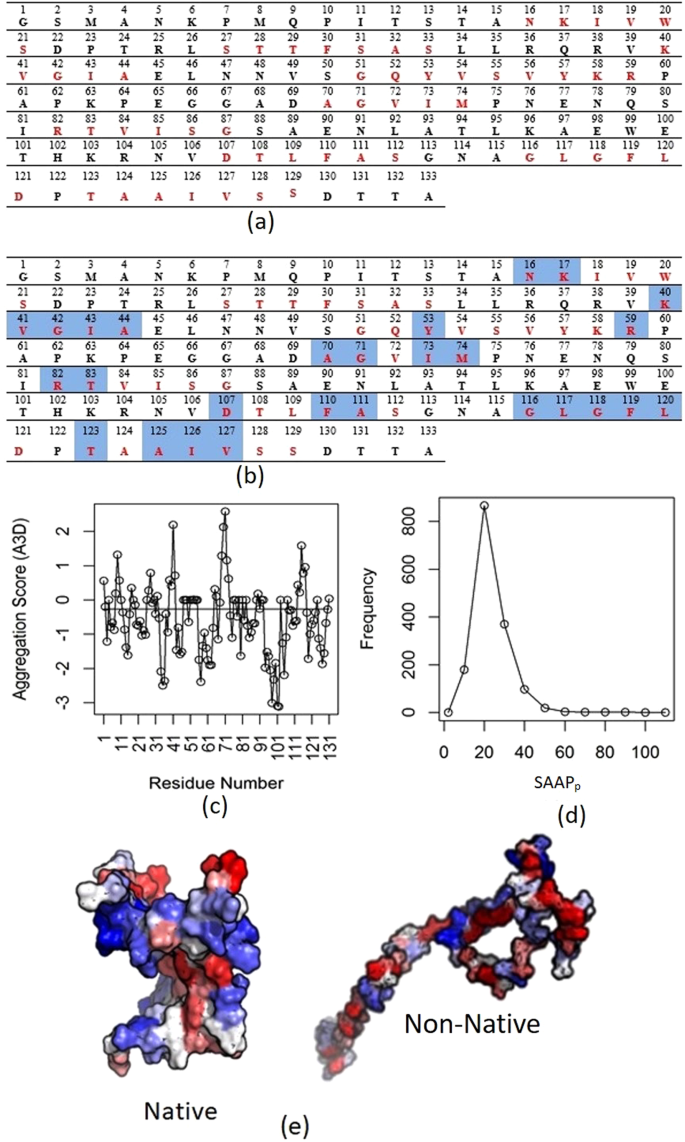
Figure 2 from Aggrescan3D (A3D) 2.0: prediction and engineering of protein solubility | Semantic Scholar

Computational prediction of protein aggregation: Advances in proteomics, conformation-specific algorithms and biotechnological applications - ScienceDirect

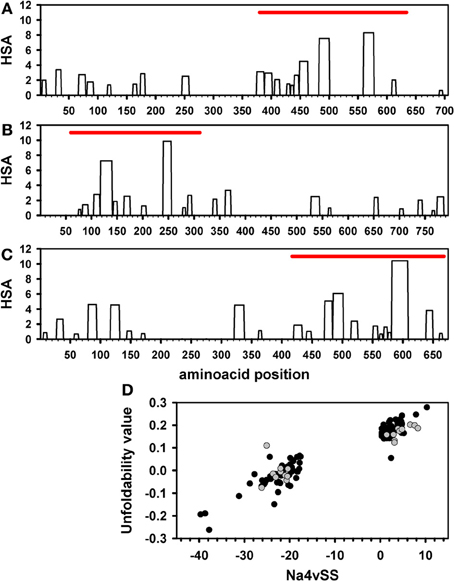
AggScore: Prediction of aggregation‐prone regions in proteins based on the distribution of surface patches - Sankar - 2018 - Proteins: Structure, Function, and Bioinformatics - Wiley Online Library
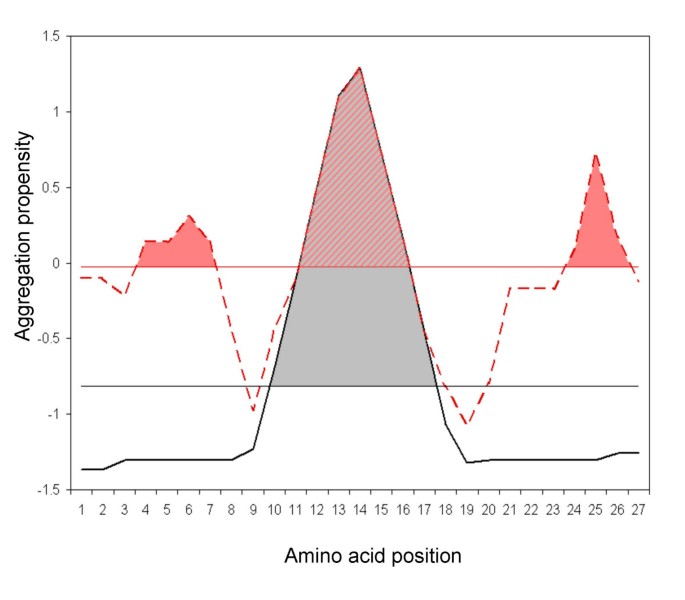
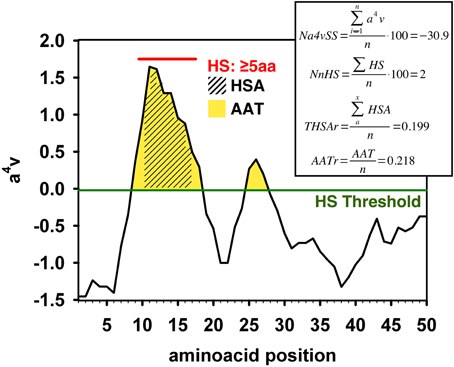
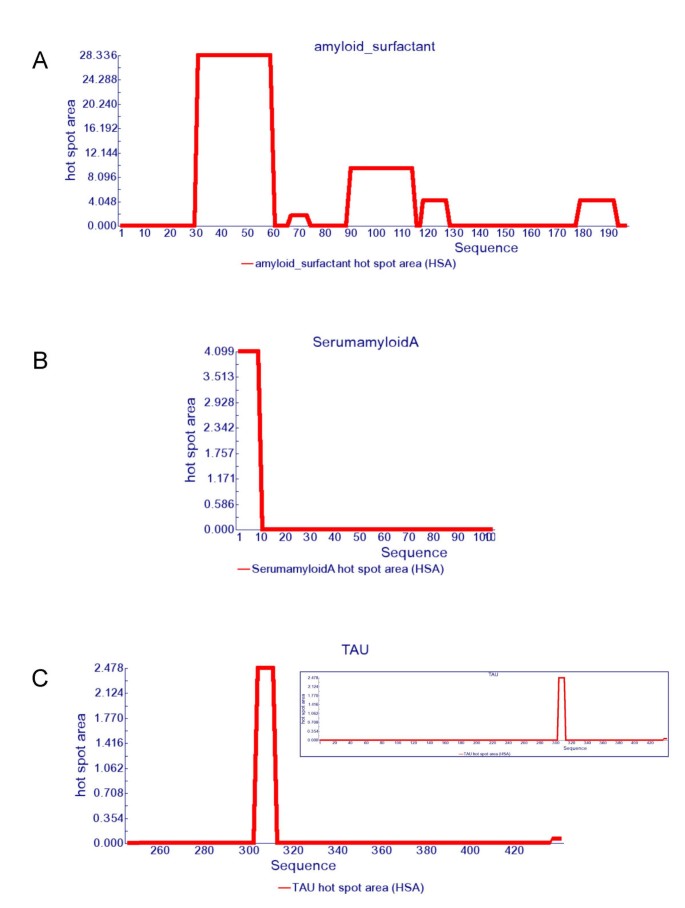
AGGRESCAN: a server for the prediction and evaluation of "hot spots" of aggregation in polypeptides | BMC Bioinformatics | Full Text
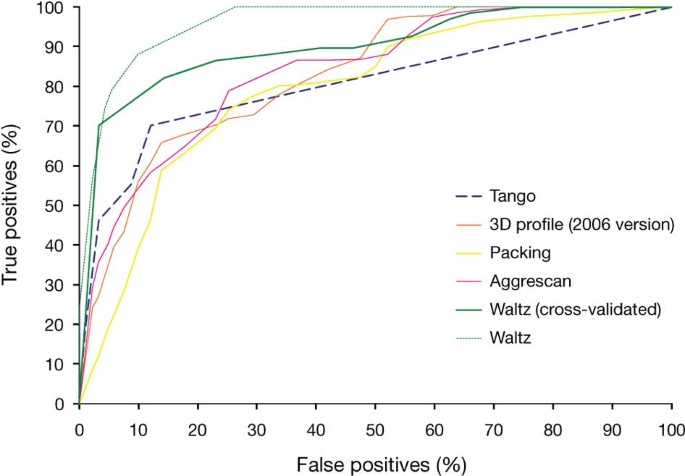
Addendum: Exploring the sequence determinants of amyloid structure using position-specific scoring matrices | Nature Methods
![PDF] AGGRESCAN3D (A3D): server for prediction of aggregation properties of protein structures | Semantic Scholar PDF] AGGRESCAN3D (A3D): server for prediction of aggregation properties of protein structures | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/bc8d0acd077eed82b977fbf9d706415235ef1fe6/7-Figure4-1.png)
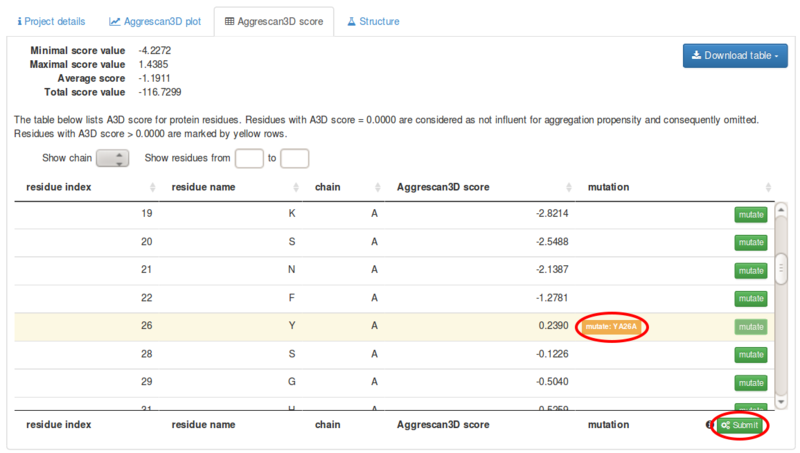
PDF] AGGRESCAN3D (A3D): server for prediction of aggregation properties of protein structures | Semantic Scholar

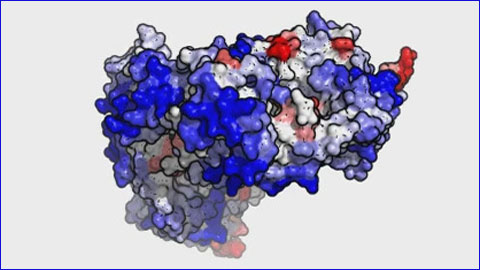
Figure 1 from AGGRESCAN3D (A3D): server for prediction of aggregation properties of protein structures | Semantic Scholar
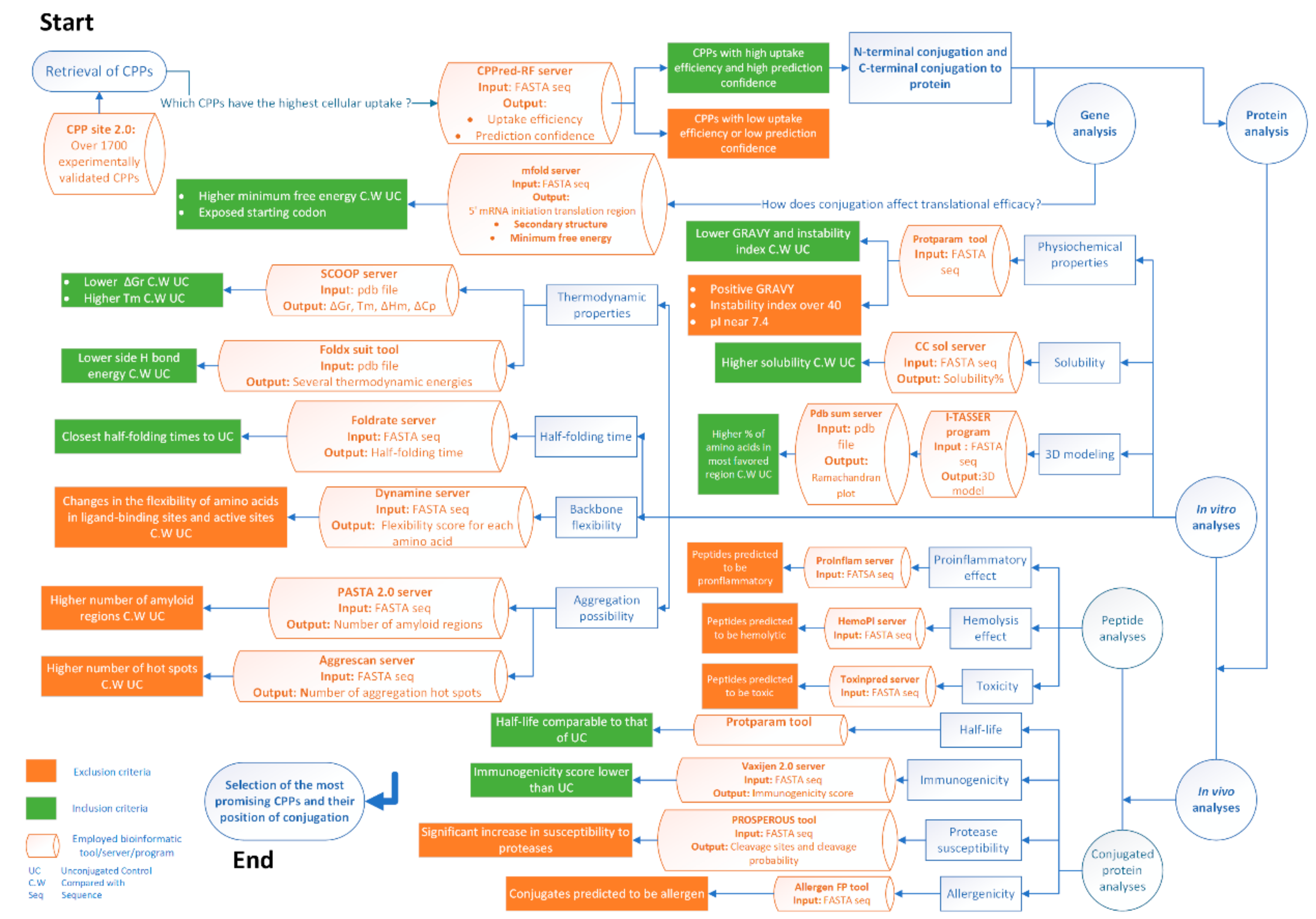
Molecules | Free Full-Text | Considerations on the Rational Design of Covalently Conjugated Cell-Penetrating Peptides (CPPs) for Intracellular Delivery of Proteins: A Guide to CPP Selection Using Glucarpidase as the Model Cargo